
The aim of this post is to do a small MCMC comparison of Stan and NumPyro/Jax (naming here inspired by the GNU/Linux Richard Stallman copypasta) on the CPU. The comparison is purely about performance, and does not address things like how easy they are to use etc. Keep in mind that such a comparison is hard to do well and the implementations here might not be optimal.
Stan is a probabilistic programming language, which uses its own math library for automatic differentiation. So it includes everything from the modeling language and inference algorithms to the library that computes gradients that the algorithms need. NumPyro on the other hand, is "a lightweight probabilistic programming library that provides a NumPy backend for Pyro. We rely on JAX for automatic differentiation and JIT compilation to GPU / CPU" as they write on their website. And to make things even more confusing, Pyro is built ontop of PyTorch.
Here I use the same one-compartment PK model as in this post, but now it is a Bayesian population PK model, so there is a population mean for each parameter and then subject-specific deviation from it.
functions {
real conc_one(real t, real dose, real ka, real cl, real vc) {
real k = cl / vc;
real comp = ka / (ka - k) * (exp(-k * t) - exp(-ka * t));
real conc_mg_per_L = (dose / vc) * comp;
return conc_mg_per_L * 1000;
}
}
data {
int<lower=1> S;
int<lower=1> N;
array[N] int<lower=1, upper=S> subj;
array[N] real<lower=0> time_hr;
int<lower=1> n_doses;
array[n_doses] real<lower=0> dose_time;
array[n_doses] real<lower=0> dose_mg;
array[N] real<lower=0> conc_obs;
}
parameters {
real mu_log_cl;
real mu_log_vc;
real mu_log_ka;
real<lower=0> sigma_log_cl;
real<lower=0> sigma_log_vc;
real<lower=0> sigma_log_ka;
vector[S] log_cl_raw;
vector[S] log_vc_raw;
vector[S] log_ka_raw;
real<lower=1e-3> sigma_obs;
}
transformed parameters {
vector[S] log_cl = mu_log_cl + sigma_log_cl * log_cl_raw;
vector[S] log_vc = mu_log_vc + sigma_log_vc * log_vc_raw;
vector[S] log_ka = mu_log_ka + sigma_log_ka * log_ka_raw;
}
model {
mu_log_cl ~ normal(log(10), 0.4);
mu_log_vc ~ normal(log(20), 0.4);
mu_log_ka ~ normal(-2, 0.4);
sigma_log_cl ~ normal(0, 0.3);
sigma_log_vc ~ normal(0, 0.3);
sigma_log_ka ~ normal(0, 0.3);
log_cl_raw ~ std_normal();
log_vc_raw ~ std_normal();
log_ka_raw ~ std_normal();
sigma_obs ~ normal(0, 0.4);
vector[S] cl = exp(log_cl);
vector[S] vc = exp(log_vc);
vector[S] ka = exp(log_ka);
for (n in 1:N) {
int s = subj[n];
real conc_hat = 0;
for (d in 1:n_doses) {
if (time_hr[n] >= dose_time[d]) {
real t_rel = time_hr[n] - dose_time[d];
conc_hat += conc_one(t_rel, dose_mg[d], ka[s], cl[s], vc[s]);
}
}
conc_hat = fmax(conc_hat, 1e-7);
conc_obs[n] ~ lognormal(log(conc_hat), sigma_obs);
}
}
from __future__ import annotations
import math
import jax
# Enable 64-bit precision to match stan
jax.config.update("jax_enable_x64", True)
import jax.numpy as jnp
import numpyro
import numpyro.distributions as dist
numpyro.set_platform("cpu")
def pk_one_comp(
time_hr: jnp.ndarray,
dose_mg: jnp.ndarray,
ka: jnp.ndarray,
cl: jnp.ndarray,
vc: jnp.ndarray,
) -> jnp.ndarray:
"""Closed-form one-compartment oral solution (ng/mL)."""
k = cl / vc
comp = (ka / (ka - k)) * (jnp.exp(-k * time_hr) - jnp.exp(-ka * time_hr))
conc_mg_per_L = (dose_mg / vc) * comp
conc = conc_mg_per_L * 1000.0
return jnp.clip(conc, a_min=1e-7, a_max=None)
def _one_comp_population_model(
subject_idx: jnp.ndarray,
time_hr: jnp.ndarray,
dose_mg: jnp.ndarray,
conc_obs: jnp.ndarray,
n_subjects: int,
conc_fn,
):
"""Shared population model body; conc_fn picks closed-form or ODE solver."""
mu_log_cl = numpyro.sample("mu_log_cl", dist.Normal(math.log(10.0), 0.4))
mu_log_vc = numpyro.sample("mu_log_vc", dist.Normal(math.log(20.0), 0.4))
mu_log_ka = numpyro.sample("mu_log_ka", dist.Normal(-2.0, 0.4))
sigma_log_cl = numpyro.sample("sigma_log_cl", dist.TruncatedNormal(0.0, 0.3, low=0.0))
sigma_log_vc = numpyro.sample("sigma_log_vc", dist.TruncatedNormal(0.0, 0.3, low=0.0))
sigma_log_ka = numpyro.sample("sigma_log_ka", dist.TruncatedNormal(0.0, 0.3, low=0.0))
with numpyro.plate("subjects", n_subjects):
log_cl = numpyro.sample("log_cl", dist.Normal(mu_log_cl, sigma_log_cl))
log_vc = numpyro.sample("log_vc", dist.Normal(mu_log_vc, sigma_log_vc))
log_ka = numpyro.sample("log_ka", dist.Normal(mu_log_ka, sigma_log_ka))
cl = jnp.exp(log_cl)
vc = jnp.exp(log_vc)
ka = jnp.exp(log_ka)
sigma_obs = numpyro.sample("sigma_obs", dist.TruncatedNormal(0.0, 0.4, low=1e-3))
conc_hat = conc_fn(time_hr, dose_mg, ka[subject_idx], cl[subject_idx], vc[subject_idx])
numpyro.sample("obs", dist.LogNormal(jnp.log(conc_hat), sigma_obs), obs=conc_obs)
def pop_pk_model_one_comp(
subject_idx: jnp.ndarray,
time_hr: jnp.ndarray,
dose_mg: jnp.ndarray,
conc_obs: jnp.ndarray,
n_subjects: int,
):
"""Population model using the closed-form PK solution."""
return _one_comp_population_model(subject_idx, time_hr, dose_mg, conc_obs, n_subjects, pk_one_comp)
The NumPyro NUTS is initialized to the below values and timed after a dummy run. This is to ensure that the JIT compilation is not included in the runtime.
init_values = {
"log_cl": jnp.array(math.log(10.0)),
"log_vc": jnp.array(math.log(20.0)),
"log_ka": jnp.array(-2.0),
"sigma_log_cl": jnp.array(0.1),
"sigma_log_vc": jnp.array(0.1),
"sigma_log_ka": jnp.array(0.1),
"log_cl_subj": jnp.full((data["n_subjects"],), math.log(10.0)),
"log_vc_subj": jnp.full((data["n_subjects"],), math.log(20.0)),
"log_ka_subj": jnp.full((data["n_subjects"],), -2.0),
"sigma_obs": jnp.array(0.4),
}
def init_from_values(init_values, site):
if site["type"] == "sample" and not site["is_observed"]:
val = init_values.get(site["name"])
if val is not None:
return val
kernel = NUTS(model, target_accept_prob=0.95, init_strategy=partial(init_from_values, init_values))
kwargs = {
"time_hr": jnp.array(data["obs_time"]),
"dose_mg": {"dose_times": jnp.array(data["dose_times"]), "dose_amounts": jnp.array(data["dose_amounts"])},
"conc_obs": jnp.array(data["obs_conc"]),
}
if "obs_subject_idx" in data and "n_subjects" in data:
kwargs["subject_idx"] = jnp.array(data["obs_subject_idx"])
kwargs["n_subjects"] = data["n_subjects"]
# Trigger compilation with a tiny run (not timed)
warmup_dummy, samples_dummy = 1, 1
mcmc_dummy = MCMC(
kernel,
num_warmup=warmup_dummy,
num_samples=samples_dummy,
num_chains=2,
chain_method="sequential",
progress_bar=False,
)
mcmc_dummy.run(rng_key, **kwargs)
# Timed run
mcmc = MCMC(
kernel,
num_warmup=warmup,
num_samples=samples,
num_chains=2,
chain_method="sequential",
progress_bar=True,
)
start = time.perf_counter()
mcmc.run(random.split(rng_key)[0], **kwargs)
runtime = time.perf_counter() - start
Also the Stan NUTS sampler is initialized similarly and a warmup run is done to ensure that the model is compiled before timing.
# Warm run to avoid counting compilation/initialization overhead
warm_seed = int(np.random.default_rng().integers(0, 2**31 - 1))
init_median = {
"mu_log_cl": math.log(10.0),
"mu_log_vc": math.log(20.0),
"mu_log_ka": -2.0,
"sigma_log_cl": 0.1,
"sigma_log_vc": 0.1,
"sigma_log_ka": 0.1,
"log_cl_raw": np.zeros(stan_data["S"]),
"log_vc_raw": np.zeros(stan_data["S"]),
"log_ka_raw": np.zeros(stan_data["S"]),
"sigma_obs": 0.4,
}
model.sample(
data=stan_data,
iter_warmup=1,
iter_sampling=1,
chains=1,
parallel_chains=1,
show_progress=False,
seed=warm_seed,
fixed_param=True,
adapt_engaged=False,
inits=init_median,
)
start = time.perf_counter()
run_seed = int(np.random.default_rng().integers(0, 2**31 - 1))
fit = model.sample(
data=stan_data,
iter_warmup=warmup,
iter_sampling=samples,
chains=2,
parallel_chains=1,
show_progress=True,
adapt_delta=0.95,
inits=init_median,
seed=run_seed,
)
runtime = time.perf_counter() - start
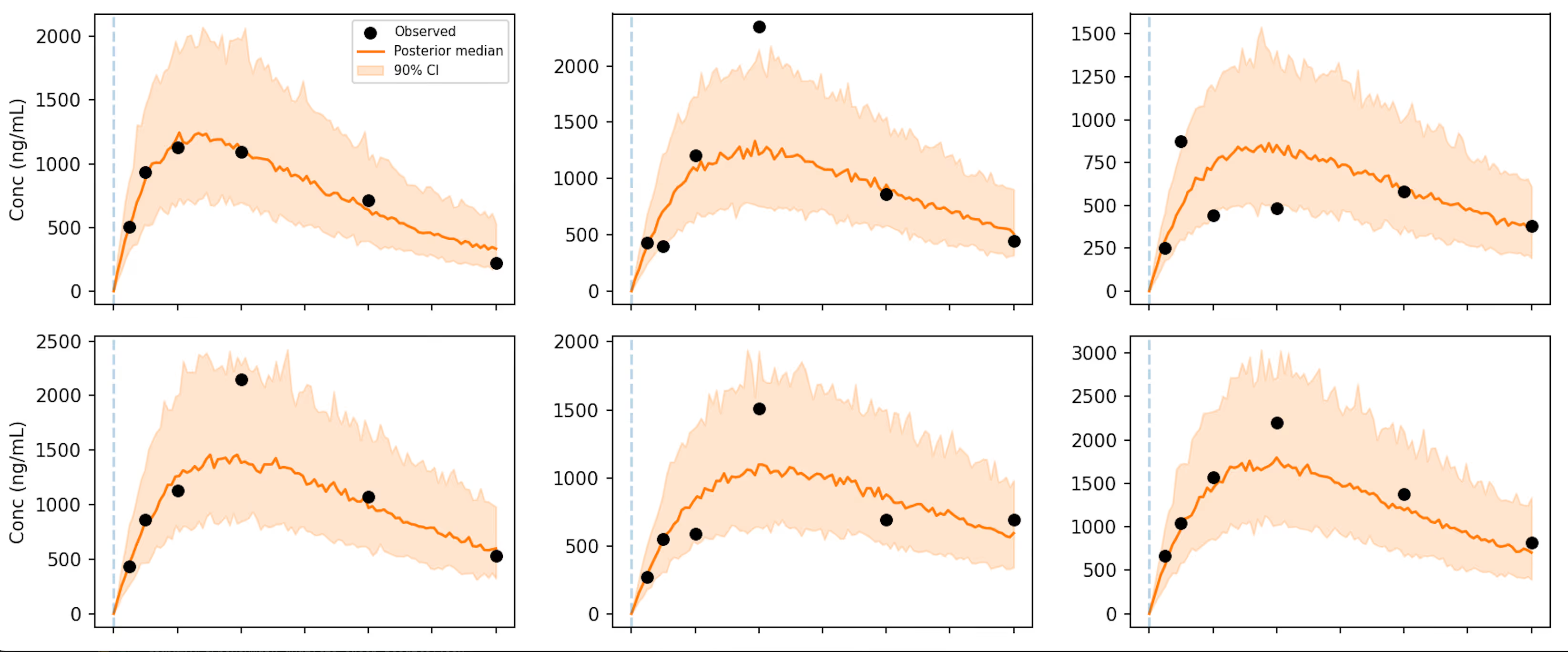
I simulate data with just a single dose for each subject, with the same 100 mg dose for all subjects. The below plot gives an idea of what a simulated data set looks like (6 example subjects over 12 hours).
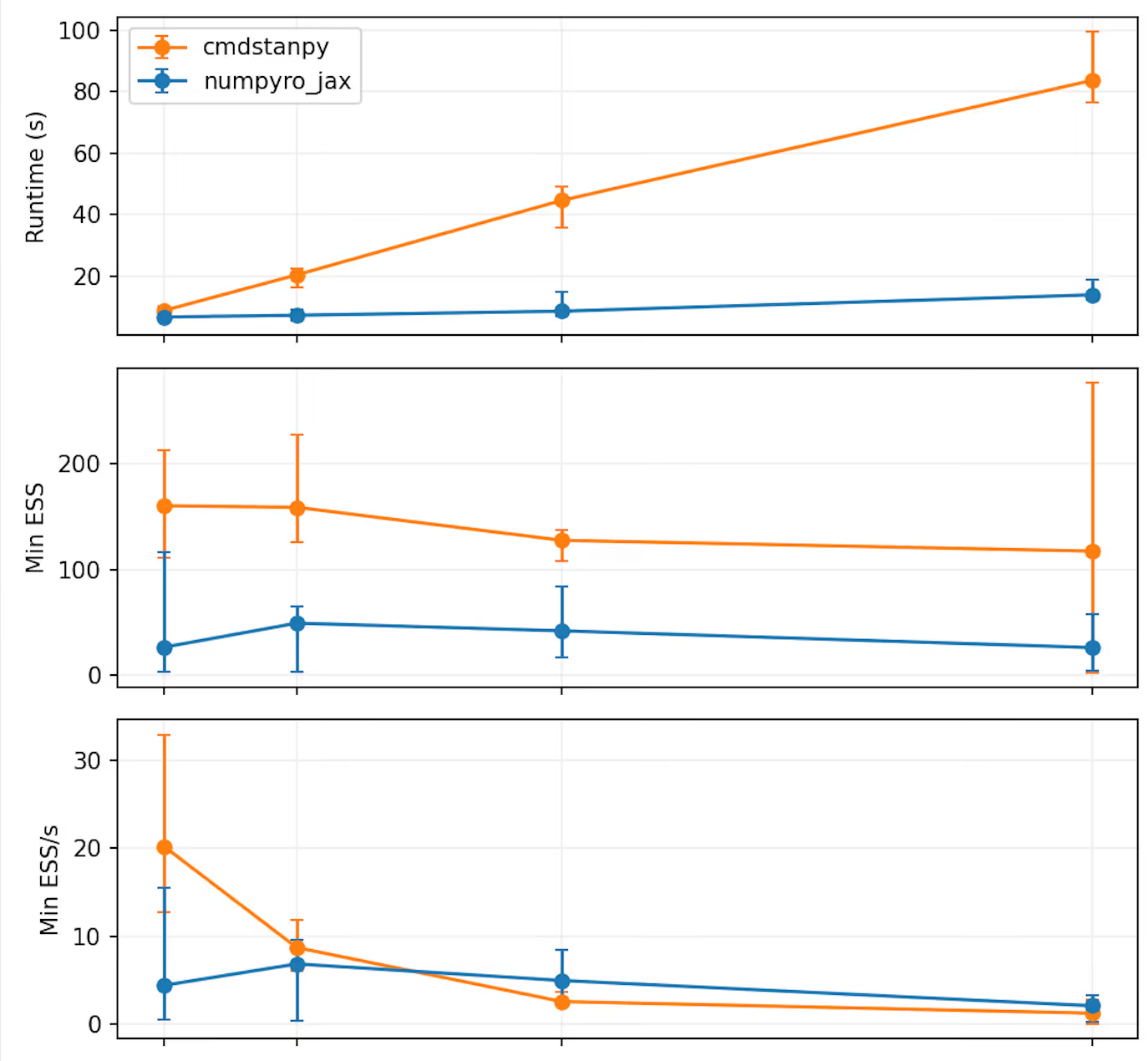
Two sequential chains (1000/1000 warmup/sampling iterations) are run for each simulated data set. The number of subjects is varied from 16 to 256. The experiment is repeated 5 times. The draws are transformed into arviz InferenceData objects and arviz is then used to estimate the effective sample size (ESS) as
ess = az.ess(idata, method="bulk")
Median results over the 5 repeats are shown below. The "errorbars" extend to the minimum and maximum values.
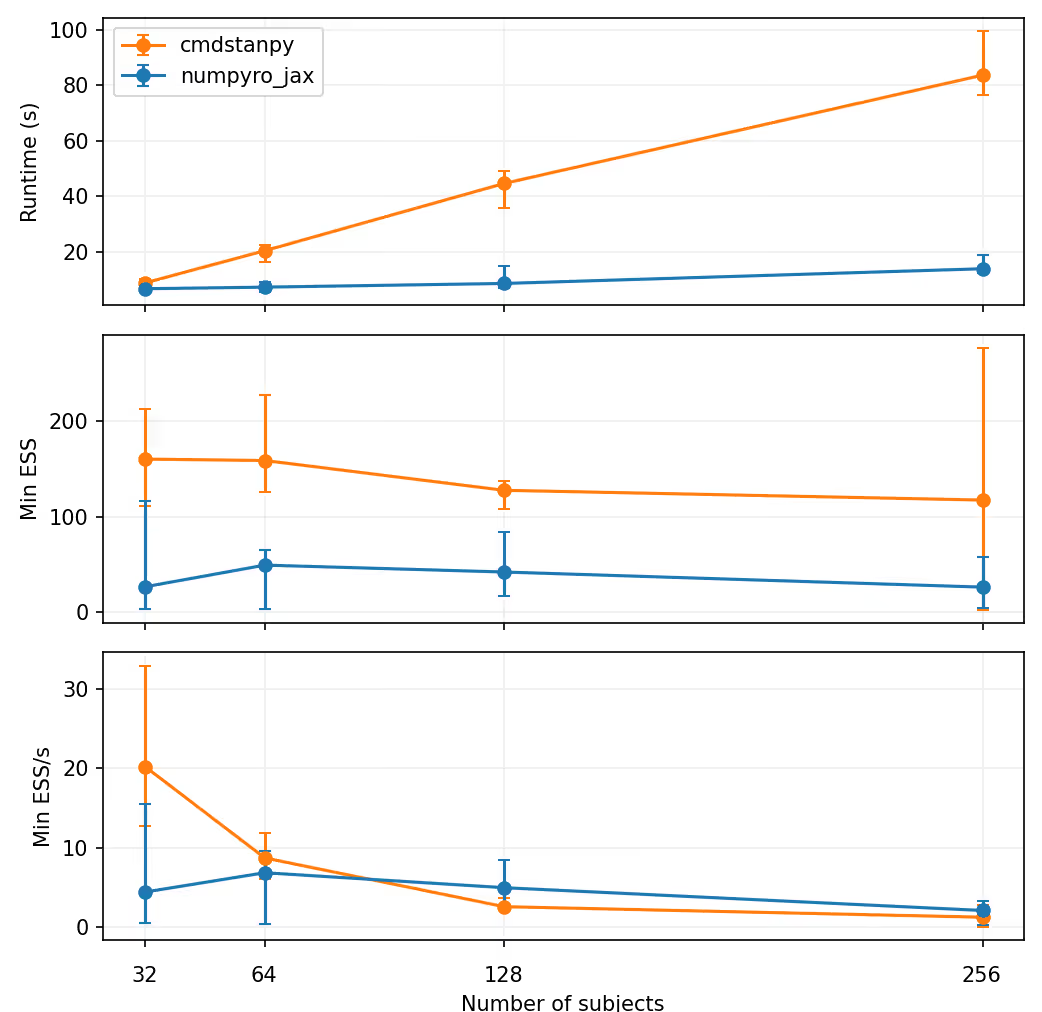
It is clear that NumPyro does run faster, and the runtime seems to increase somewhat linearly as a function of the number of subjects. Somehow NumPyro it consistently seems to get a lower ESS, though. Whether this difference is due to some small differences in the NUTS implementations or something else, is not clear to me.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.

Weighing pros and cons of NCA vs. PK modeling.

This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.