
I was recently talking to several cardiovascular researchers who are running a trial for patients receiving anticoagulant medication. The trial results are not out yet, but they wanted to understand what to expect from prior work in this area and sent me the paper titled “Effects of oral anticoagulation in people with atrial fibrillation after spontaneous intracranial haemorrhage (COCROACH): prospective, individual participant data meta-analysis of randomised trials.” (Al-Shahi Salman et al. 2023)
It just so happens that I am developing a tool, codenamed Orion, for semi-automated (at first, may be fully automated later) data extraction and Bayesian meta-analysis (BMA) that is geared towards evidence synthesis from published literature. It is already pretty good at auto-object detection (tables and figures) and structured extraction, but I have not yet wired in the BMA, which I hope to do by running Stan in the browser using Brian Ward’s Wasm-based implementation.
The main summary from the paper is the forest plot below.
.avif)
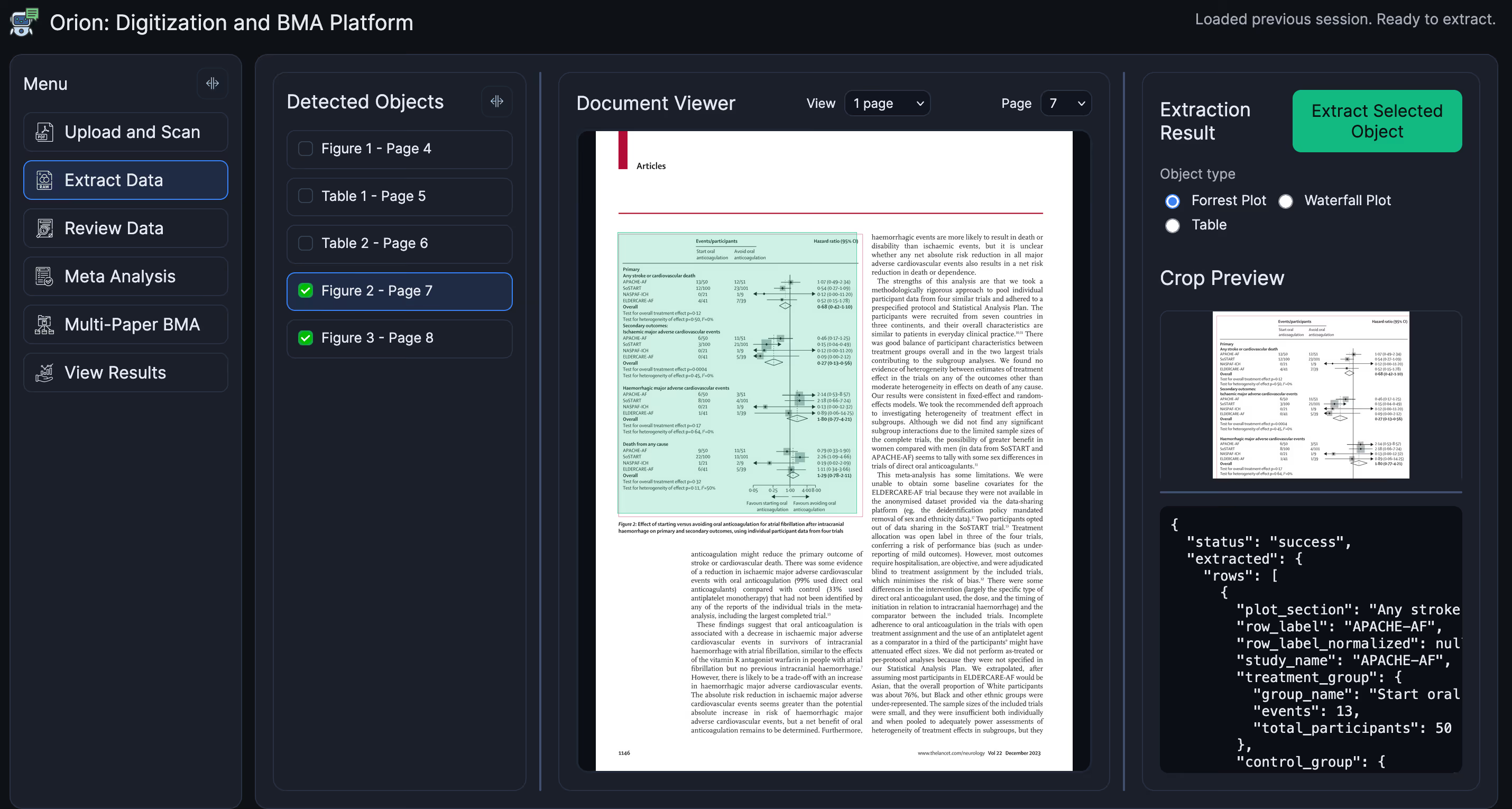
We will use the data from this forest plot to get posterior probabilities of the treatment effects. The following screenshot demonstrates that Orion (correctly) detected three figures and four tables. I clicked on Figure 2; Orion then extracted the data using Gemini Pro into a structured JSON format (snippet in the lower right). For full transparency, object and boundary detection is done using a combination of Surya, pdf2image + PyMuPDF, and PyTorch.
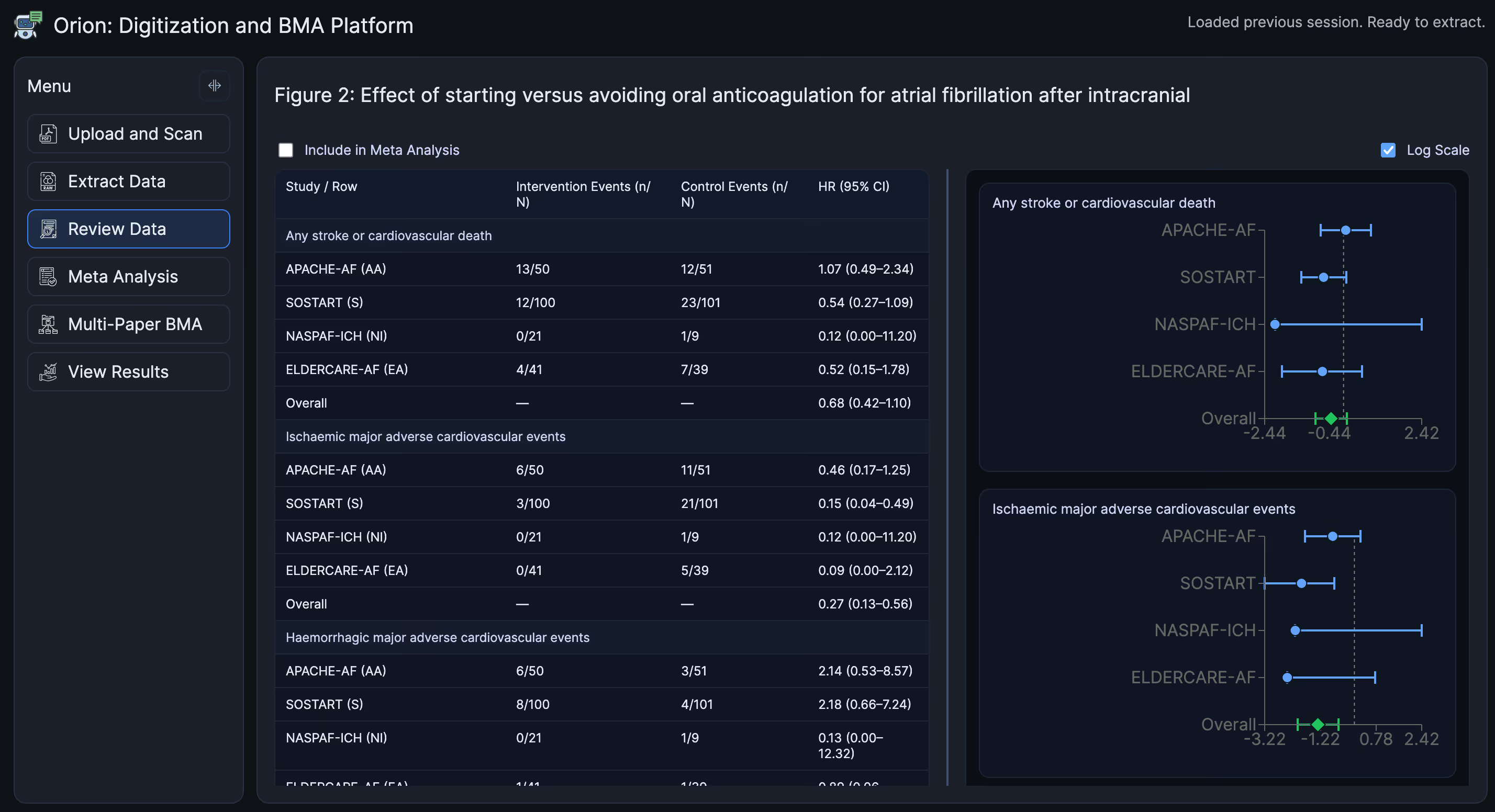
Once the structured data is extracted it can be displayed and downloaded in CSV and JSON. The table below shows the extracted data and the reproduced frequentist confidence intervals.
We fit a binomial model for treatment effects offline; that model is described next.
As always, there are several ways to parameterize this type of model. The Stan User’s Guide has an example using the log odds and standard errors, which can be computed from data, but it would not work in this case, since one of the trials had zero events. Instead, we use a direct binomial parameterization with a logit transform for the probabilities.
To be clear, we will run four separate meta-analyses, since we are tracking four separate clinical endpoints. When we work with patient-level data, we develop much more realistic models where there is dependency among different end points, say a minor bleeding event and intracranial haemorrhage.
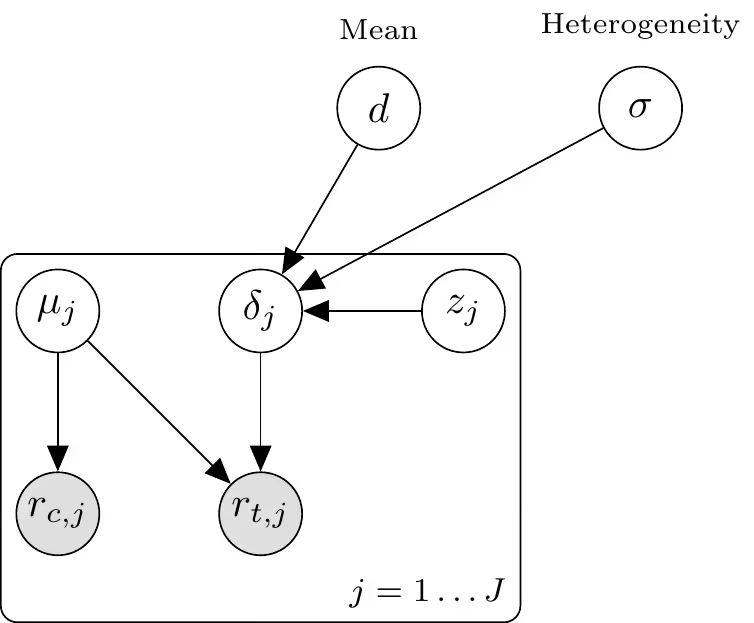
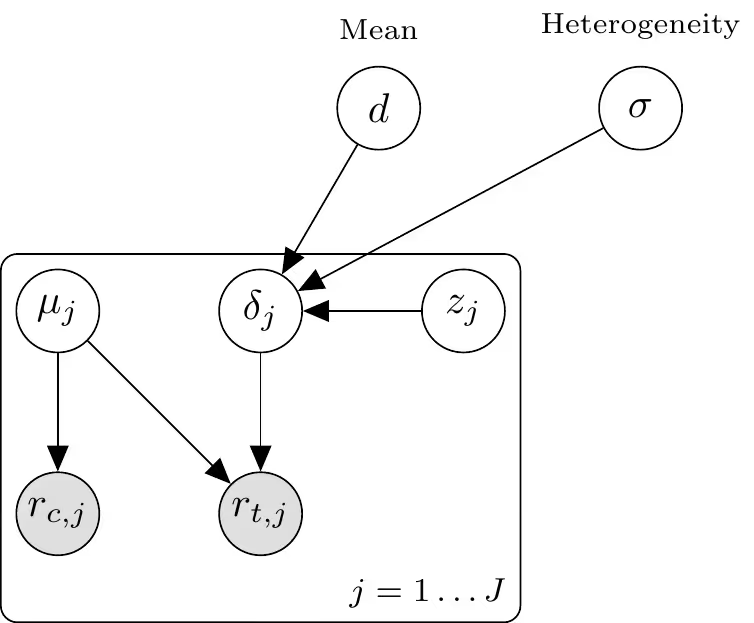
What follows is a description of the meta-analysis model we apply separately to each of the four endpoints. Our data consists of trials. For each trial $j=1, \ldots, J$, let $r_{c, j}$ and $r_{t, j}$ denote the number of events in the control and treatment arms, respectively, out of $n_{c, j}$ and $n_{t, j}$ participants.
We model the event counts using a Binomial likelihood with a logit link function:
$$\begin{aligned} r_{c, j} & \sim \operatorname{Binomial}\left(n_{c, j}, p_{c, j}\right) \\ r_{t, j} & \sim \operatorname{Binomial}\left(n_{t, j}, p_{t, j}\right) \\ \operatorname{logit}\left(p_{c, j}\right) & =\mu_{j} \\ \operatorname{logit}\left(p_{t, j}\right) & =\mu_{j}+\delta_{j}\end{aligned}$$
where $\mu_{j}$ represents the study-specific baseline log-odds of the event, and $\delta_{j}$ represents the study-specific log-odds ratio (treatment effect).
The study-specific effects $\delta_{j}$ are assumed to be exchangeable and drawn from a normal population distribution with mean $d$ and standard deviation $\sigma$. To improve sampling efficiency (non-centered parameterization), we define:
$$\begin{aligned} \delta_{j} & =d+\sigma \cdot z_{\delta, j} \\ z_{\delta, j} & \sim \mathcal{N}(0,1)\end{aligned}$$
This implies the marginal distribution $\delta_{j} \sim \mathcal{N}\left(d, \sigma^{2}\right)$.
We assign weakly informative priors to the hyperparameters:
$$\begin{aligned} \mu_{j} & \sim \mathcal{N}\left(-2,2^{2}\right) \\ d & \sim \mathcal{N}\left(0,1^{2}\right) \\ \sigma & \sim \mathcal{N}^{+}\left(0,1^{2}\right)\end{aligned}$$
The parameters are transformed back to the Odds Ratio (OR) scale for interpretation:
Note that the treatment effect $\mathrm{OR}_{\text {mean }}$ is not adequate for predicting the effect in a new study, because it ignores population heterogeneity. That is captured by $\mathrm{OR}_{\text{pred}}$.
The following plate diagram may help to visualize the hierarchical parameter-data structure.
It is worth noting that Bayesian and frequentist estimates are not an apples-to-apples comparison: the paper reports hazard ratios from Cox regression, while we use a binomial model and report odds ratios. We do not emphasize that distinction here.
The above model is implemented in Stan. One of many great things about Stan is how closely it matches the mathematical definition.
data {
int<lower=1> J; // num of trials
array[J] int<lower=0> r_c; // num events, control
array[J] int<lower=0> r_t; // num events, treatment
array[J] int<lower=1> n_c; // num cases, control
array[J] int<lower=1> n_t; // num cases, treatment
}
parameters {
vector[J] mu; // baseline log-odds
real d; // population mean effect
real<lower=0> sigma; // population heterogeneity
vector[J] z_delta; // non-centering
}
transformed parameters {
vector[J] delta = d + sigma * z_delta;
}
model {
mu ~ normal(-2, 2);
d ~ std_normal();
sigma ~ std_normal();
z_delta ~ std_normal();
r_c ~ binomial_logit(n_c, mu);
r_t ~ binomial_logit(n_t, mu + delta);
}
generated quantities {
vector[J] study_OR = exp(delta);
real mean_OR = exp(d);
real pred_OR = exp(normal_rng(d, sigma));
}
We perform full Bayesian inference using cmdstanr and compute the relevant estimands, which are $\mathrm{Pr}(\mathrm{OR}_{\mathrm{mean}} < 1)$ for each clinical endpoint.
These are directionally consistent with the known effects of anticoagulants, which reduce the probability of ischaemic events at the cost of increasing bleeding (haemorrhagic) risk.
Finally, we can compare the frequentist and Bayesian estimates, even though, as mentioned before, they come from different models.
.avif)
You can clearly see the pooling effect from smaller studies but otherwise fairly close agreement. Of course, the Bayesian approach yields more directly interpretable probabilities for the treatment effect and estimates of population heterogeneity.
If you want to reproduce the analysis, the data are available here. The R script used to run the analysis can be found here.
Al-Shahi Salman, Rustam, Jacqueline Stephen, Jayne F Tierney, Steff C Lewis, David E Newby, Adrian R Parry-Jones, Philip M White, et al. 2023. “Effects of Oral Anticoagulation in People with Atrial Fibrillation After Spontaneous Intracranial Haemorrhage (COCROACH): Prospective, Individual Participant Data Meta-Analysis of Randomised Trials.” The Lancet Neurology 22 (12): 1140–49. https://doi.org/10.1016/s1474-4422(23)00315-0.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
From R/Stan to Julia/Turing

This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.