
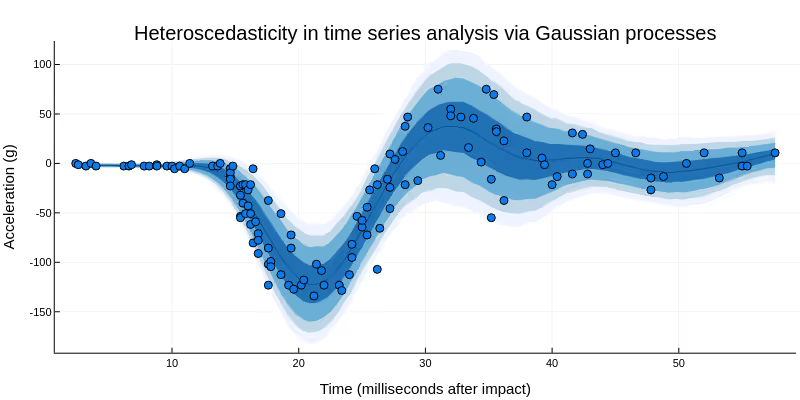
Nonparametric statistical model components are a flexible tool for imposing structure on observable or latent processes. Usually, nonparametric statistical models have an uncountably infinite number of free parameters, but reduce their effective flexibility by constraining these parameters to covary in ways that guard against overfitting.
Gaussian processes represent one family of such models. Mathematically, putting a Gaussian process prior with mean function $\mu$ and covariance function $k$ on a function $f$, i.e.
$$f \sim \mathrm{GP}(\mu, k),$$
implies that for any $x_1$ and $x_2$, the joint prior distribution of $f(x_1)$ and $f(x_2)$ is a multivariate Gaussian distribution with mean $[\mu(x_1), \mu(x_2)]^T$ and covariance $k(x_1, x_2)$. The mean function $\mu$ can be as simple as a constant value (e.g. $\mu(x) = 0$), but often depends on other model parameters and covariates. Common choices for the covariance function $k$ include:
For an interactive visualization of different combinations of kernel functions, hyperparameters, and observations, see Ti John’s excellent Interactive Gaussian process Visualization.
By wrapping the function $f$ in appropriate link functions, which may in turn depend on other model parameters or covariates, Gaussian processes allow the modeler both to "let the data speak for itself" and to impose prior beliefs on the modelled function $f$. For example:
Every Gaussian process comes with a set of "tunable" or inferrable hyperparameters. In the field of machine learning, these hyperparameters are usually optimized and then fixed for further inference. While this approach often works very well in the "big data" setting typical of machine learning, with health data, we are often left with less data than we would like, but still have to reach robust conclusions and make high-stakes decisions. To do so, we must properly account for any relevant residual uncertainty, which regularly includes the choice of hyperparameters for the covariance functions $k$.
For a function $f \sim \mathrm{GP}(\mu, k)$ depending on a single covariate $x$, and with the covariance function $k$ being the squared exponential function, there are two positive hyperparameters:
Without previous experience, it can be exceedingly difficult to specify principled priors for these hyperparameters. Poorly chosen priors can, in turn, substantially affect final conclusions and decisions.
Even after settling on a covariance function and hyperpriors, there remains the issue of scaling.
For exact Gaussian process regression with variable hyperparameters, computing the prior density has cubic complexity in the number of distinct observation points. This means that evaluating the prior or posterior density even for the simplest possible model $y \sim \mathrm{Normal}(\mu(x), \sigma^2)$ with $\mu \sim \mathrm{GP}(0, k)$ will take 8 times as long if you double the number of distinct values of the covariate $x$. This happens because, under the hood, to evaluate the prior density of $\mu(x) \sim \mathrm{Normal}(0, \Sigma)$ for $n$ distinct covariate values $x$, we have to first construct the full covariance matrix $\Sigma$ and then perform its Cholesky decomposition, which has the cubic complexity in $n$.
Scaling quickly becomes prohibitive if the hyperparameters are allowed to vary - which as discussed previously is often a prerequisite to making robust decisions under uncertainty. After all, "full" Bayesian inference, as realized through the use of Markov Chain Monte Carlo methods, often requires evaluating the posterior log density and its gradient many thousands to millions of times. Even fixing the hyperparameters will only reduce the scaling from cubic to effectively quadratic, because while fixed hyperparameters allow the Cholesky decomposition to be precomputed and reused, we will still have to multiply with or divide by it at every posterior log density evaluation.
Fortunately, there are solutions, which however come with their own challenges.
There are two main ways to discretize Gaussian processes, both of which use a finite number $m$ of basis functions $\phi_i$ to approximate a function $f \sim \mathrm{GP}(0, k)$ as
$$f(x) \approx \sum_{i=1}^m w_i \phi_i(x):$$
Both approximations eliminate the at least quadratic complexity in the number $n$ of distinct covariate values $x$, if the number $m$ of basis functions does not depend on $n$:
The main benefit of the inducing points approximation over the Hilbert space approximation is due to the locality of the basis functions of the inducing point approximation: If you only need to approximate your Gaussian process in small and potentially disjointed subsets of its underlying covariate space, the inducing point approximation allows you to only allocate basis functions to these subsets, which is something that the Hilbert space approximation is incapable of, as its basis functions are fully "global". This can be particularly noteworthy in higher-dimensional covariate spaces, where the "full" Hilbert space approximation always scales exponentially with the dimension, while the inducing points approximation potentially does not. In practice however, this advantage seldomly materializes, and there are good alternatives to the "full" Hilbert space approximation such as additive Gaussian processes as implemented in Juho's `lgpr` package.
Generally, because we want to propagate the uncertainty about the hyperparameters and thus will not fix them, we are using the Hilbert space Gaussian process approximation at Generable. However, even though the scaling of this approximation is linear and thus optimal when considering only the cost of evaluating a single posterior log density, making use of it in practice can often be quite challenging. Specifically, it is common to observe much worse than linear scaling in the number of observations if the approximation is used naively. The root of this problem lies in the oft-quoted specter of "bad" or "inappropriate" parametrizations, and does also affect both the exact Gaussian process regression and its approximation via inducing points.
We will revisit this problem and its solution in a later post in this series.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.

Weighing pros and cons of NCA vs. PK modeling.

This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.