
Cell-free DNA (cfDNA) utilization is growing in cancer treatment and drug development. Its use in cancer genetic profiling has become increasingly routine, and is now also offering sensitive disease measurement through minimal residual disease (MRD) monitoring. This approach has shown particular clinical promise when combined with neoadjuvant therapy. Beyond MRD applications, cfDNA is emerging as a valuable pharmacodynamic marker. The BR.36 trial validates this potential, examining whether cfDNA can replace radiographic RECIST scans. Early results are encouraging, demonstrating stronger correlation with clinical endpoints than conventional radiographic measurements.
Today I want to illustrate how cfDNA provides a unique tool for early detection of disease changes, using results from clinical samples. To do this, I will use an example of early diagnosis with cell-free DNA from some work we published in Nature Medicine. I will use examples from this paper below to illustrate the concept, however the work covers a lot more than I will reference, so I encourage you to read the paper if you are curious about the genomics of Richter syndrome [1].
Richter syndrome (RS) is a lymphoma that is associated with the progression of chronic lymphocytic leukemia (CLL), which is a cancer of the B cell. Patients with RS have a poor overall survival, typically less than one year. Traditionally diagnosed by a pathology morphologic assessment, it carries a misdiagnosis rate of as high as 20%.
The unique strength of cfDNA is that it carries molecular signals from all lesions in the body, not just the primary disease, so it has prognostic value even in heme malignancies like CLL. In another study we worked on, a comparison of cfDNA to multiple biopsies showed how local heterogeneity is well captured in the blood plasma. (Parikh et al.)
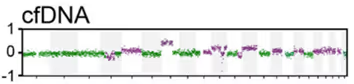
In Parry et al., we compared whole exome sequencing from whole blood to plasma (from the same vial) and a lymph-node biopsy. Whole exome sequencing of peripheral blood mononuclear cells (PBMCs) revealed only CLL variants, even after Richter syndrome diagnosis. At the same time, sequencing of cfDNA in the blood plasma detects RS variants even earlier than a clinical diagnosis was made.
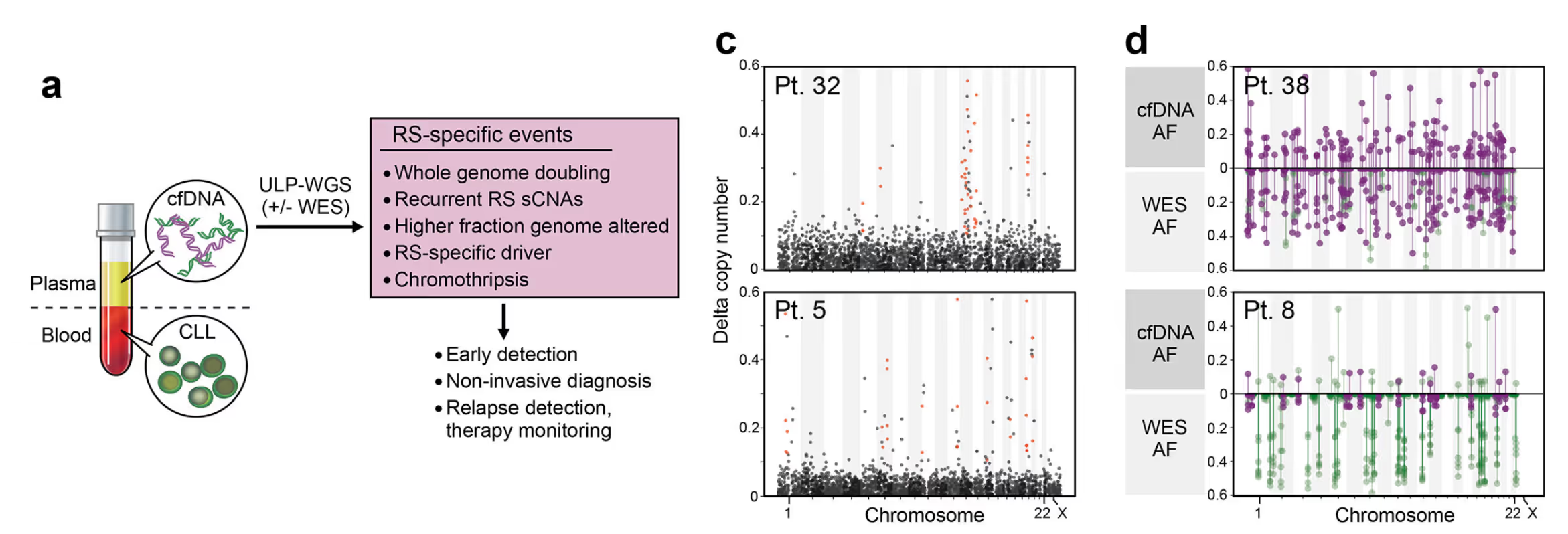
This is most clear when looking at contemporaneous samples from one patient that reveal the amount of genomic information missed when not utilizing cfDNA. To illustrate this lets look at some sequencing results. While CLL is typically characterized by very few mutations, RS is typically associated with large copy number events and mutations. Lets compare two samples from the same patient, comparing the profile of CLL with the profile of the associated RS progression.
Note the large scale copy number changes present on the top plot, completely absent on the bottom. This difference in mutations highlights the high heterogeneity within a patient and underscores the importance of looking at cfDNA.
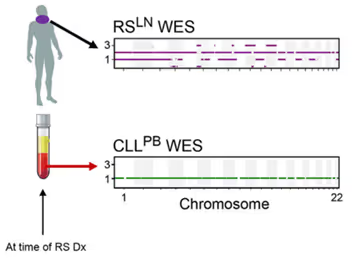
If we look at the cfDNA collected at the same exact time as the two samples above, you see the lymph node variants present, but this time measured directly from the blood plasma.
This signal allows cfDNA to not only measure heterogeneous mechanisms resistance as we saw in Parikh et al. but also provide early diagnosis. For patients where blood was available early, here is a comparison of the plasma sequencing months before diagnosis with the eventual biopsy:
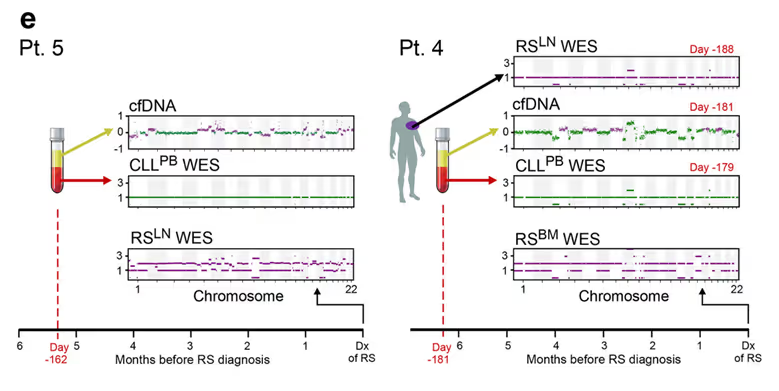
For both of these patients, even 5+ months before diagnosis, the variants of Richter’s syndrome are clearly detectable in cfDNA. For a patient over the course of two years after alloSCT, a therapy where allogenic stem cells are transplanted, the emergence of a relapse is measured months before it is diagnosed and the success of treatment is measured soon after therapy is started.
In addition to making early diagnosis possible, the sequencing information collected can be used to infer clinically important factors of the disease. For RS, in patients where the disease is a progression of the existing leukemia the mutations associated with it are found in addition to the variants shared by the leukemia cells.
Utilizing a tool we built, clonal heterogeneity analysis can be used to reconstruct the disease lesions from these sequencing samples [3]. You can think of these results similarly to what you could measure with single cell sequencing - individual cell populations and lesions can be identified.
Here is an example “tree” structure, with the RS mutations outlined in purple.
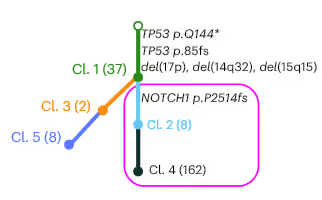
In this case mutations in TP53 are shared by all CLL cells (Cl. 1). The RS biopsy contains a NOTCH1 frameshift mutation in addition to the shared Cl. 1 mutations (TP53). That NOTCH1 mutation is not found in the PBMCs.
However in some patients, the disease in the lymph nodes was found to be unrelated to the established mutations in the CLL. For instance, in this patient, none of the mutations from the blood cells (Cl. 1, Cl. 3, Cl. 5) were found in the biopsy.
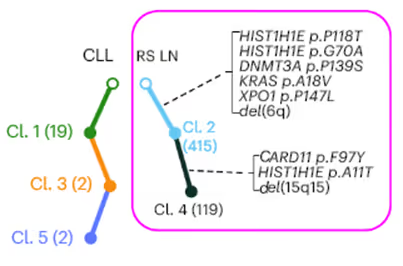
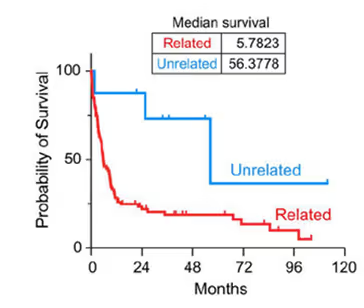
This analysis has direct clinical implication - in Parry et al. we saw that patients with de novo unrelated lymph node disease had far better survival than patients who’s CLL had progressed directly.
[1] Parry, E. M., Leshchiner, I., Guièze, R., et al. (2023). Evolutionary history of transformation from chronic lymphocytic leukemia to Richter syndrome. Nature Medicine, 29(1), 158–169.
[2] Parikh, A. R., Leshchiner, I., Elagina, L., et al. (2019). Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nature Medicine, 25(9), 1415–1421.
[3] Leshchiner, I., Mroz, E. A., Cha, et al. (2023). Inferring early genetic progression in cancers with unobtainable premalignant disease. Nature Cancer, 4(4), 550–563.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.

Weighing pros and cons of NCA vs. PK modeling.

This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.