
Model selection is a difficult process, particularly in high dimensional settings, dependent observations, and sparse data regimes. In this post, I will discuss a common misconception about selecting models based on values of the objective function generated from optimization algorithms in sparse data settings. TL;DR Don’t do it.
As always, I will start by simulating some data.
n <- 10s
set.seed(1)
x <- rnorm(n)
a <- 1
b <- 2
y <- a + b * x + rnorm(n, sd = 3)
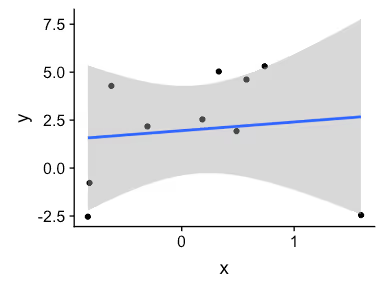
qplot(x, y) + geom_smooth(method = lm)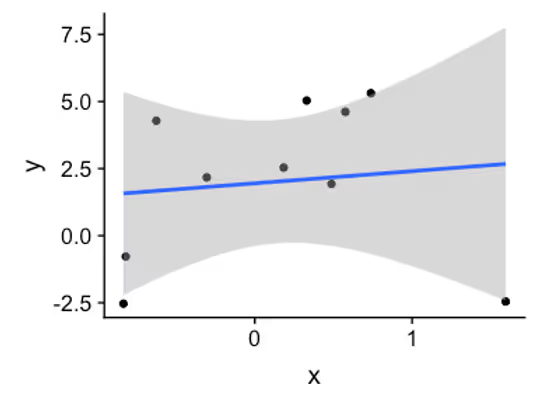
In order to fit this model with maximum likelihood, we are going to need the normal log probability density function (LPDF) and the objective function – negative of the LPDF evaluated and accumulated at each data point $y$. The normal PDF is defined as follows:
$$f(y \mid \mu, \sigma) = \frac{1}{\sqrt{2\pi \sigma}} \exp\left( -\frac{1}{2} \left( \frac{y - \mu}{\sigma} \right)^2 \right)$$
Taking the logs, yields:
$$f_{\log}(y \mid \mu, \sigma) = \log\left( \frac{1}{\sqrt{2\pi \sigma}} \right) - \frac{1}{2} \left( \frac{y - \mu}{\sigma} \right)^2$$
In R, this function can be defined as follows:
normal_lpdf <- function(y, mu, sigma) {
log(1 / (sqrt(2*pi) * sigma)) - 1/2 * ((y - mu) / sigma)^2
}
Computationally, the constants should be pre-computed, but we do not bother with this here. For sanity check, we compare to built-in R’s dnorm().
dnorm(c(0, 1), mean = 0, sd = 2, log = TRUE)
## [1] -1.612086 -1.737086
normal_lpdf(c(0, 1), mu = 0, sigma = 2)
## [1] -1.612086 -1.737086
The objective function that we will minimize is the sum of the negative log likelihood evaluated at each data point y.
# For comparison, MLE estimate using lm() function
arm::display(lm(y ~ x))
## lm(formula = y ~ x)
## coef.est coef.se
## (Intercept) 1.95 1.01
## x 0.45 1.35
## ---
## n = 10, k = 2
## residual sd = 3.15, R-Squared = 0.01
# Negative log likelihood
log_lik <- function(theta, x, y) {
ll <- -sum(normal_lpdf(y,
mu = theta[1] + theta[2] * x,
sigma = theta[3]))
return(ll)
}
# MLE estimate using custom log likelihood
res <- optim(par = rep(1, 3), # initial values
fn = log_lik,
x = x,
y = y)
paste("Estimated value of a:", round(res$par[1], 2))
## [1] "Estimated value of a: 1.95"
paste("Estimated value of b:", round(res$par[2], 2))
## [1] "Estimated value of b: 0.45"
paste("Estimated value of sigma:", round(res$par[3], 2))
## [1] "Estimated value of sigma: 2.82"
paste("Objective function value:", round(res$value, 2))
## [1] "Objective function value: 24.56"
Now let’s see if we can decrease the value of the objective function by adding a quadratic term to the model.
arm::display(lm(y ~ x + I(x^2)))
## lm(formula = y ~ x + I(x^2))
## coef.est coef.se
## (Intercept) 3.88 0.82
## x 2.31 0.97
## I(x^2) -3.84 1.04
## ---
## n = 10, k = 3
## residual sd = 1.96, R-Squared = 0.67
log_lik_xsqr <- function(theta, x, y) {
ll <- -sum(normal_lpdf(y,
mu = theta[1] + theta[2]*x + theta[3]*(x^2),
sigma = theta[4]))
return(ll)
}
res_xsqr <- optim(par = rep(1, 4),
fn = log_lik_xsqr,
x = x,
y = y)
round(res_xsqr$par, 2)
## [1] 3.88 2.31 -3.84 1.64
paste("Objective function value:", round(res_xsqr$value, 2))
## [1] "Objective function value: 19.12"
Not surprisingly, the value of the objective function is lower and $R^2$ is higher.
What happens when we increase n, by say 10-fold. We still get the reduction in the objective function value but it is a lot smaller in percentage terms.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.
This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.

An exercise I probably should have done earlier.

Where more data can be bad (if you don’t handle them right)

This is a comment related to the post above. It was submitted in a form, formatted by Make, and then approved by an admin. After getting approved, it was sent to Webflow and stored in a rich text field.